“AN INTERVIEW WITH FRANCESCO RUGGERI, JUNIOR RESEARCH FELLOW DEPARTMENT OF CHEMISTRY and CENTER FOR MISFOLDING DISEASES, UNIVERSITY OF CAMBRIDGE"

Francesco Simone Ruggeri is a Junior Research fellow at the Darwin College and Research Fellow at the Department of Chemistry Center for Misfolding Diseases at the University of Cambridge. Both at Cambridge and previously at École Polytechnique Fédérale de Lausanne (EPFL),his research is focused on the biophysical characterization of proteins at the single molecule scale by means of atomic force microscopy based techniques. This approach has brought new insights into the formation and structural characterization of misfolding proteins and their correlation with the onset of neurodegenerative disorders
The Park AFM is fundamental for my research on amyloids because of some extremely important features: i) it has an extremely low electrical noise in the order of 25 pm, which enable high-resolution measurements at the nanoscale, ii) the decoupled XY and Z scanners, which enable easier post-processing (flattening) of the image at these small scales, together with a consistent exclusion of the molecules during the 3-D AFM map flattening, and iii) the versatility of the instrument interface and software make the tool extremely easy to use and allow high-throughput, highresolution measurements.
3-D Printing as a technological feat that started as hobbyists’innovative way of building things has growninto a wellaccepted additive manufacturing (AM) technology. It has now penetrated invarious manufacturing industries such as in electronics manufacturing,bioengineering, food fabrication, and in almost anything we can think of.
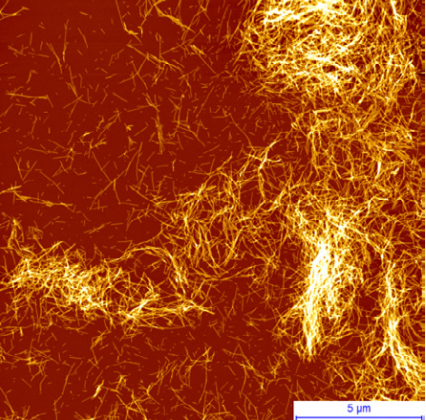
Today, more than 40 million people worldwide are affected by dementia and neurodegenerative disorders (Ruggeri, Habchiet al. 2016). Onset of these and more than fifty other human diseases is associated at the molecular level with insoluble fibrillar protein aggregates, termed amyloids (Fig. 1).
Figure 1. AFM image of amyloid fibrils of α-synuclein, the protein involved in the onset of Parkinson’s disease
The objective of my research is to address the challenge of understanding the role amyloids serve in the onset of human diseases by studying aggregates one at the time to achieve a molecular level characterization of amyloid properties and formation. Indeed, the high complexity in the connection between protein aggregation and cellular toxicity may be reconciled by taking into account a structural diversity and heterogeneity of amyloid aggregates formed. Therefore, there is need for tools able to characterize the biophysical properties of heterogeneous amyloids at the nanoscale in vitro, in relation to the molecular mechanism of onset of the pathologies and to pharmacological treatments in vivo. My approach is to study amyloid heterogeneity from a biophysical point of view and to develop and apply novel methodologies to shed light on the physical determinants of protein aggregation.
More specifically, I pursue this objective by bringing together atomic force microscopy, with cutting-edge biophysical techniques, such as microfluidics and biosensors to protein science and biophysics. As consequence, a major part of my work is the successful application of nanoscale imaging by means of atomic force microscopy (AFM) in order to investigate the biophysical properties and formation of amyloids at the single aggregate scale. AFM has emerged in the last decade as one of the most powerful and versatile single molecule techniques because of the possibility to acquire 3-dimensional morphology maps of specimens on a surface. This capability has been widely used in the field of protein aggregation and amyloid fibril formation. Indeed, a simple AFM map provides extremely valuable information at the nanometer scale on the structure of amyloid fibrils, such as height, width, periodicity, and the packing of single protofilaments inside mature fibrils (Ruggeri, Habchi et al. 2016).
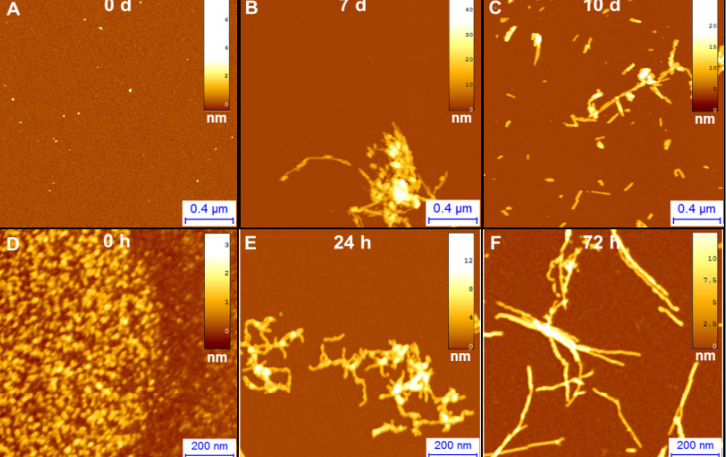
A single molecule technique, such as AFM, allows one to gain information on the fibrillization process and on individual protein structural transition. The possibility to analyze the morphology at several time points, during the process of amyloid aggregation, enables one to shed light on the mechanisms of protein misfolding, on the pathway of aggregation and the hierarchical polymorphic process of assembly. Figure 2 displays the fibrillization process of α-synuclein and Aβ42 by means of AFM imaging after deposition of the proteins on a positively functionalize APTES-mica substrate (Ruggeri, Adamcik et al. 2015).
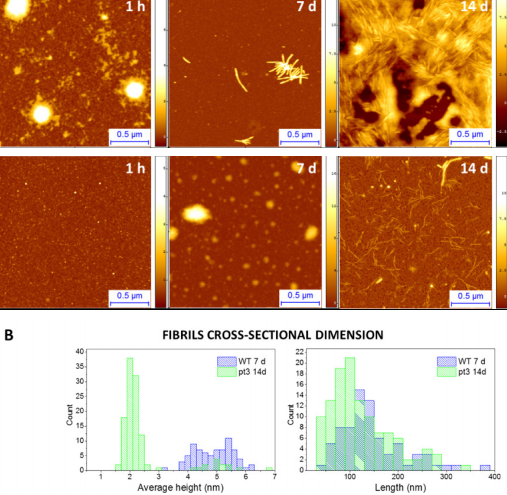
In addition, AFM imaging can be also used to compare the kinetics of aggregation of different mutated forms of the same protein. Indeed, the study of the effects of mutation on the aggregation of amyloidogenic proteins is of fundamental importance to shed light on their role in both physiology and association with disease. For example, the aggregation of Huntingtin protein and its mutated forms is linked to the onset of Huntington’s disease. Increasing evidence suggests that specific N-terminal post-translation modifications within the N-terminus of Huntingtin play a role in the pathogenesis of the illness. For this reason, we focused on the study of their fibrillation and on differences between wild type and a mutated form of this protein (Ansaloni, Wang et al. 2014, Ruggeri, Vieweg et al. 2016, Chiki, DeGuire et al. 2017). As an example, we show here how our AFM studies allowed us to quantitatively follow the kinetics of amyloid fibril formation, and to show that the N-terminus phosphorylated (pT3) mutated form of the protein significantly slows the fibrillation process by forming fibrils with smaller diameters and shorter lengths (Fig. 3)
Figure 2.AFM imaging of A)α-synuclein, involved in Parkinson‘s and B) Aβ42, involved in Alzheimer‘s fibrillization process after incubation of the proteins for 0 d, 7 d, 10 d.
Figure 3.(A) Aggregation of wild type and mutated pT3 huntingtin followed by AFM. (B) Height and length distributions of mutated pT3 (green) a wild type oligomeric structures
References
Ansaloni, A., Z. M. Wang, J. S. Jeong, F. S. Ruggeri, G. Dietler and H. A. Lashuel (2014). "One-pot semisynthesis of exon 1 of the Huntingtin protein: New tools for elucidating the role of posttranslational modifications in the pathogenesis of Huntington's disease." Angewandte Chemie - International Edition53(7): 1928-1933.
Chiki, A., S. M. DeGuire, F. S. Ruggeri, D. Sanfelice, A. Ansaloni, Z.-M. Wang, U. Cendrowska, R. Burai, S. Vieweg, A. Pastore, G. Dietler and H. A. Lashuel (2017). "Mutant Exon1 Huntingtin Aggregation is Regulated by T3 Phosphorylation-Induced Structural Changes and Crosstalk between T3 Phosphorylation and Acetylation at K6." Angewandte Chemie129(19): 5286-5291. Ruggeri, F. S., J. Adamcik, J. S. Jeong, H. A. Lashuel, R. Mezzenga and G. Dietler (2015). "Influence of the β-Sheet Content on the Mechanical Properties of Aggregates during Amyloid Fibrillization." Angewandte Chemie - International Edition.
Ruggeri, F. S., J. Habchi, A. Cerreta and G. Dietler (2016). "AFM-Based Single Molecule Techniques: Unraveling the Amyloid Pathogenic Species." Curr Pharm Des22(26): 3950-3970.
Ruggeri, F. S., S. Vieweg, U. Cendrowska, G. Longo, A. Chiki, H. A. Lashuel and G. Dietler (2016). "Nanoscale studies link amyloid maturity with polyglutamine diseases onset." Sci Rep6: 31155.